Coding in R is useless without interesting research questions; and even the best questions remain unanswered without data. RStudio provides a number of convenient ways to acces data, among which the possibility to write SQL code chunks in Rmarkdown, to run these chunks and to assign the value of the query result directly to a variable of your choice. No such thing is available yet for SPARQL queries; the ones that alow you to navigate gigantic knowledge graphs that incarnate the conscience of the semantic web. This is where the SPARQLchunks package comes in.
- It allows you to run SPARQL chunks in Rmarkdown files.
- It provides inline functions to send a SPARQL queries to a user-defined endpoint and retrieve data in _dataframe_ form (`sparql2df`) or _list_ form (`sparql2list`).
Endpoints can be reached from behind corporate firewalls thanks to automatic proxy detection.
Installation
Most users can install by running this command:
remotes::install_github("aourednik/SPARQLchunks", build_vignettes = TRUE)Code language: R (r)If you are behind a corporate firewall on a Windows machine, direct access to GitHub might be blocked from R and RStudio. If that is your case, run this installation code instead:
proxy_url <- curl::ie_get_proxy_for_url("https://github.com")
httr::set_config(httr::use_proxy(proxy_url))
remotes::install_url("https://github.com/aourednik/SPARQLchunks/archive/refs/heads/master.zip", build_vignettes = TRUE)Code language: R (r)Use
To use the full potential of the package you need to load the library and _tell knitr that a SPARQL engine exists_:
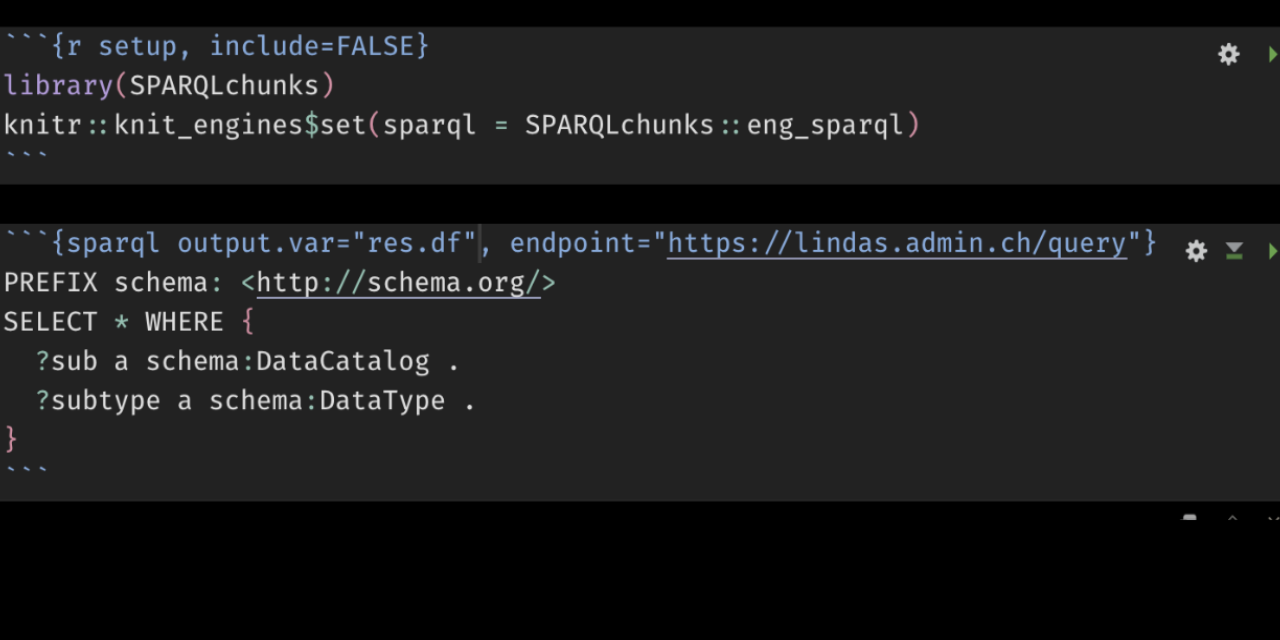
```{r setup}
library(SPARQLchunks)
knitr::knit_engines$set(sparql =SPARQLchunks::eng_sparql)
```Code language: Markdown (markdown)Once you have done so, you can run SPARQL chunks:
Chunks
Retrieve a DataFrame
output.var: the name of the data.frame you want to store the results in
endpoint: the URL of the SPARQL endpoint
autoproxy: whether or not try to use the automatic proxy detection
Example 1 (Swiss administration endpoint)
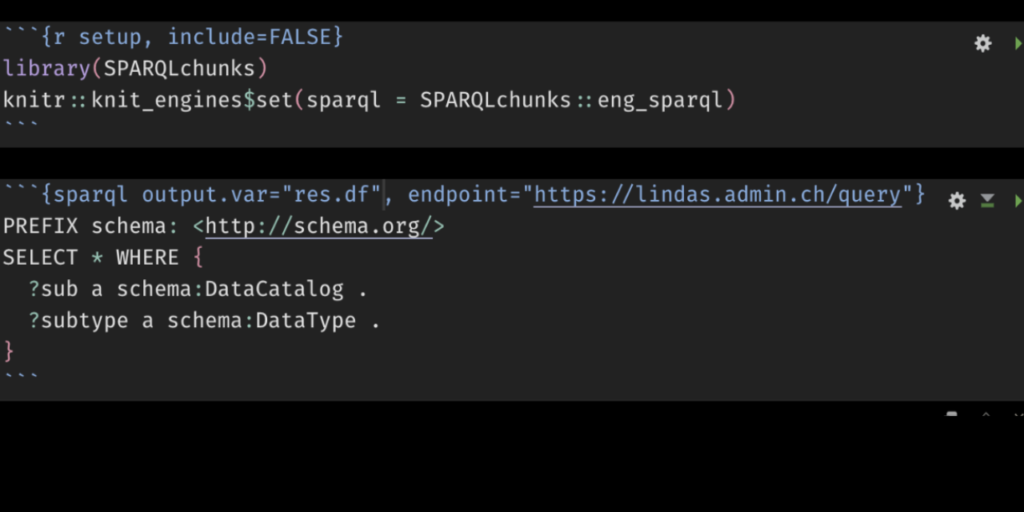
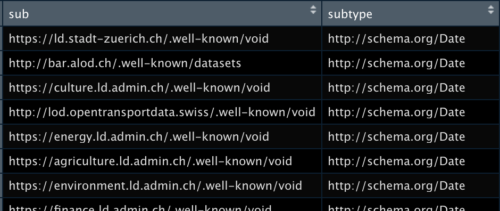
```{sparql output.var="queryres_csv", endpoint="https://lindas.admin.ch/query"}
PREFIX schema: <http://schema.org/>
SELECT * WHERE {
?sub a schema:DataCatalog .
?subtype a schema:DataType .
}
```Code language: Markdown (markdown)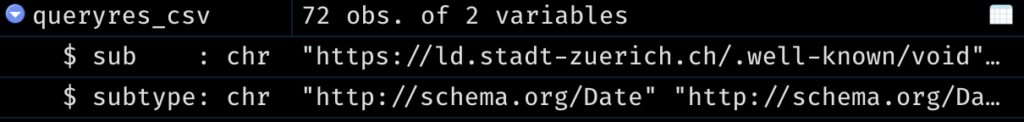
Example 2 (Uniprot endpoint)
Note the use of automatic proxy detection with autoproxy=TRUE.
```{sparql output.var="tes5", endpoint="https://sparql.uniprot.org/sparql", autoproxy=TRUE}
PREFIX up: <http://purl.uniprot.org/core/>
SELECT ?taxon
FROM <http://sparql.uniprot.org/taxonomy>
WHERE {
?taxon a up:Taxon .
} LIMIT 500
```Code language: Markdown (markdown)Example 3 (WikiData endpoint)
```{sparql output.var="res.df", endpoint="https://query.wikidata.org/sparql"}
SELECT DISTINCT ?item ?itemLabel ?country ?countryLabel ?linkTo ?linkToLabel
WHERE {
?item wdt:P1142 ?linkTo .
?linkTo wdt:P31 wd:Q12909644 .
VALUES ?type { wd:Q7278 wd:Q24649 }
?item wdt:P31 ?type .
?item wdt:P17 ?country .
MINUS { ?item wdt:P576 ?abolitionDate }
SERVICE wikibase:label { bd:serviceParam wikibase:language "[AUTO_LANGUAGE],en" . }
}
```Code language: Markdown (markdown)Retrieve a List
output.var: the name of the data.frame you want to store the results in
endpoint: the URL of the SPARQL endpoint
output.type : if set to “list”, retrieves a list (tree structure) instead of a data frame
```{sparql output.var="queryres_list", endpoint="https://lindas.admin.ch/query", output.type="list"}
PREFIX schema: <http://schema.org/>
SELECT * WHERE {
?sub a schema:DataCatalog .
?subtype a schema:DataType .
}
```Code language: Markdown (markdown)
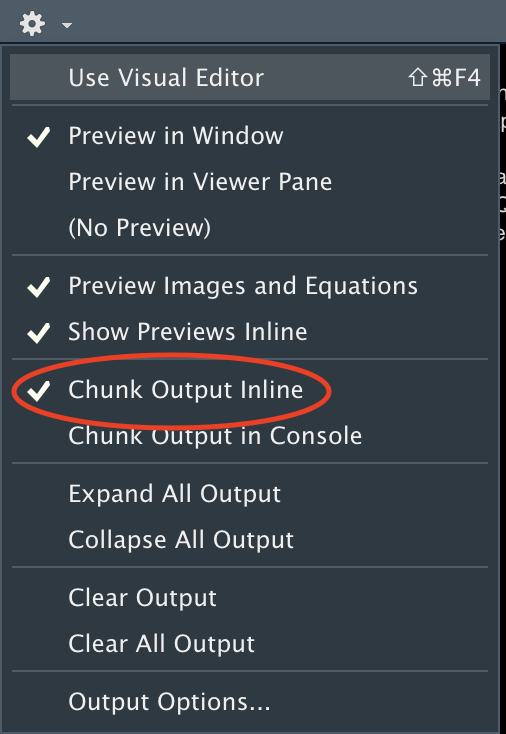
NB: Reactivate the Chunk Play Button if Needed
A bug remains in the current version of RStudio (2022.02.2) that might deactivate the “play” button for non-r chunks. To make the play button reappear for sparql chunks (as well as for sql or python chunks), make sure to select the “Chunk Output Inline” option and save your code again.
Inline code
The inline functions sparql2df and sparql2list both have the same pair of arguments: a SPARQL endpoint and a SPARQL query. Queries can be multi-line:
endpoint <- "https://lindas.admin.ch/query"
query <- "PREFIX schema: <http://schema.org/>
SELECT * WHERE {
?sub a schema:DataCatalog .
?subtype a schema:DataType .
}"Code language: R (r)Retrieve a data frame
result_df <- sparql2df(endpoint,query)Code language: R (r)The same but with attempt at automatic proxy detection:
result_df <- sparql2df(endpoint,query,autoproxy=TRUE)Code language: R (r)Retrieve a list
result_list <- sparql2list(endpoint,query)Code language: R (r)The same but with attempt at automatic proxy detection:
result_list <- sparql2list(endpoint,query,autoproxy=TRUE)Code language: R (r)Rationale
Prior to the SPARQLchunks package, there were basically two ways to access SPARQL data with R. You could have used the generic curl and httr packages, or you could have used the SPARQL package.
I recommend the curl+httr solution, which I’ve presented in an earlier version of this post before I wrapped it in a package.
The SPARQL package for R is still available on CRAN. It is a nice wrapper, but it dates back to 2013 and it doesn’t offer important parameters for HTTP(S) communication, such as setting proxies or headers, which allow you to specify the format of the returned data (XML,CSV) or to reach SPARQL endpoints from behind corporate firewalls. Also, the SPARQL package relies on RCurl, which has problems with some network settings.
As a first alternative, I’ve made an .Rmd file that allows you to execute SPARQL code chunks in RStudio. Wrapping the same functionality in an R package was an interesting exercise for me. Keep on reading if you want to understand the inner workings of the package.
Setting Up a Custom Chunk Type
One of the nice things about the knitr package is that it allows you to define custom chunk types. Let us load all necessary libraries and set up a {sparql} type.
library(httr)
library(magrittr)
library(xml2)
library(data.table)
knitr::knit_engines$set(sparql = function(options) {
code <- paste(options$code, collapse = '\n')
ep<- options$endpoint
qm <- paste(ep, "?", "query", "=", gsub("\\+", "%2B", URLencode(code, reserved = TRUE)), "", sep = "")
proxy_url <- curl::ie_get_proxy_for_url(ep)
proxy_config <- use_proxy(url=proxy_url)
varname <- options$output.var
if(is.null(options$output.type)) {
output_type <- "csv"
} else {
output_type <- options$output.type
}
if (output_type=="list") {
out <- GET(qm,proxy_config,timeout(60)) %>% read_xml() %>% as_list()
nresults <- length(out$sparql$results)
} else {
queryres_csv <- GET(qm,proxy_config,timeout(60), add_headers(c(Accept = "text/csv")))
out <- queryres_csv$content %>% rawToChar %>% textConnection %>% read.csv
nresults <- nrow(out)
}
chunkout <- ifelse(!is.null(varname),qm,out)
text <- paste("The SPARQL query returned",nresults,"results")
if (!is.null(varname)) assign(varname, out, envir = knitr::knit_global())
knitr::engine_output(options, options$code, chunkout, extra=text)
})Code language: R (r)That’s it. Just execute the chunk above and your {sparql} chunks are ready to run. Note a couple of details about the function enclosed in the definition:
The two lines proxy_url <- curl::ie_get_proxy_for_url(ep) and proxy_config <- use_proxy(url=proxy_url) use the curl package to get proxy settings of your machine. This is probably not necessary on your home computer but plays a vital role if you are trying to access the web with R from behind a corporate firewall on a Windows machine; it allows R to fetch the proxy configuration straight from your Internet Explorer.
The line queryres_csv <- GET(qm,proxy_config,timeout(60), add_headers(c(Accept = "text/csv"))) specifies that it wants the SPARQL endpoint to return a CSV instead of XML. Most modern endpoints honor the Accept = "text/csv" header. CSV’s are easy to convert to data.frames in R; XML is easier to convert to a list.
The lines varname <- options$output.var and later if (!is.null(varname)) assign(varname, out, envir = knitr::knit_global()) allow you to output the result of your SPARQL query directly to a variable storing a data.frame or, if you wish so, to a list object.
A Chunkless Approach
Nothing hinders you, of course, from using inline R script instead of chunks. First, set up your endpoint and query variables:
endpoint <- "https://lindas.admin.ch/query"
proxy_url <- curl::ie_get_proxy_for_url(endpoint)
proxy_config <- use_proxy(url=proxy_url)
query <- "PREFIX schema: <http://schema.org/>
SELECT * WHERE {
?sub a schema:DataCatalog .
?subtype a schema:DataType .
}"
querymanual <- paste(endpoint, "?", "query", "=", gsub("\\+", "%2B", URLencode(query, reserved = TRUE)), "", sep = "")Code language: R (r)Then choose your option
Get a data.frame in a Simple R Script
queryres_csv <- GET(querymanual,proxy_config, timeout(60), add_headers(c(Accept = "text/csv")))
queryres_content_csv <- queryres_csv$content %>% rawToChar %>% textConnection %>% read.csvCode language: R (r)Get a List in a Simple R Script
queryres <- GET(querymanual,proxy_config,timeout(60))
queryres_content <- queryres %>% read_xml() %>% as_list()
queryres_content$sparql$results[[1]] Code language: R (r)If your endpoint does not accept the Accept = "text/csv" header, you can also retrieve a list first and then parse it into a data.frame. Beware, though, this parsing might need adaptation for the data returned by your specific endpoint:
tableheaders <- sapply(queryres_content$sparql$head,function(x){attributes(x) %>% .$name})
table <- lapply(queryres_content$sparql$results,function(x){
d <- sapply(x,function(r){
r$uri[[1]] %>% unlist
}) %>% t %>% data.table
colnames(d) <-tableheaders
d
}) %>% rbindlist()
rm(tableheaders)Code language: R (r)